CryoEM of Inorganic Nanoparticles
Published on February 16, 2024 12:23 pm MT Updated on April 18, 2024 4:22 pm MT
Structural knowledge of atomically precise gold nanoclusters came to fruition with the first published crystal structure of Au102(pMBA)44.1 Since then, our knowledge of thiol-protected gold nanoclusters has skyrocketed with the help of Single Crystal X-Ray Diffraction (SCXRD) analysis of new nanocluster synthesis products, ligand exchange products, and more. However, SCXRD is only robust if the nanoparticle sample is homogeneous and atomically precise. An atomically precise sample allows for a high quality crystal product that diffracts in a way that produces a high resolution crystal structure.
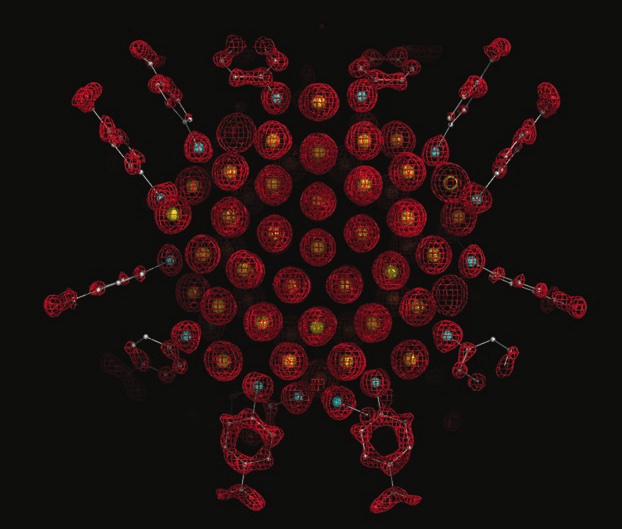
Crystal structure of Au102(pMBA)44, the first crystal structure of an atomically precise gold nanocluster.1
What if your inorganic nanoparticle (iNP) sample is not atomically precise? To date, the field is missing a routine analytical method to determine a total atomic structure (inorganic core + organic ligand shell) of a heterogeneous iNP sample. However, we can take inspiration from structural biology to develop a robust method to determine total structure of iNPs. Structural biologists routinely using cryoEM and corresponding data analysis tools to reconstruct 3D models of heterogeneous biomacromolecule samples.
CryoEM is a class of electron microscopy that involves freezing of your sample to obtain a snapshot of your sample in a layer of vitreous ice. Imaging against a background of vitreous ice as opposed to continuous carbon allows for a greater signal-to-noise ratio when imaging your sample. Our aim is to resolve the metal-ligand interface of iNPs, and imaging on a background of vitreous ice will allow us to better resolve the structure of the carbon-based organic ligands on the surface of the iNP.
We plan to utilize both Single Particle Analysis (SPA) to determine 3D structures of iNPs. SPA is a data processing method that structural biologists routinely use for developing 3D reconstructions. In general, this process utilizes automated data collection to acquire thousands of images of their sample. Then, out of these images, particles are picked and organized into 2D classes based on similarities in size, shape, orientation, etc. These classes are then used to produce a 3D electron density map of the particle, which is then refined to produce a final, high-resolution model.
As model systems, we are investigating iNPs where the organic ligands are either aligned within a repeating structural unit or confined to angular ligand pockets. This way, the conformational degrees of freedom of the ligands are reduced, allowing for increased similarities between particles that will make the reconstruction process more facile.
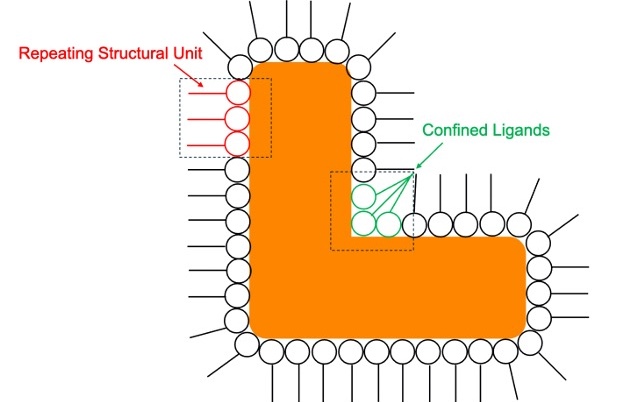
Cartoon of a nanoparticle that has a repeating structural unit of ligands along the edge of the particle (red) and ligands confined in angular ligand pockets (green).
References: Jadzinsky, P.D.; Calero, G.; Ackerson, C.J.; Bushnell, D.A.; Kornberg, R.D. Structure of a Thiol Monolayer-Protected Gold Nanoparticle at 1.1 Å Resolution. Science 2007, 318(5849), 430-433.

